Deep Learning Based Classification of Real and Synthetic Animal Images
DOI:
https://doi.org/10.58190/imiens.2025.158Keywords:
Stable Diffusion Turbo, Real–Synthetic Image Classification, MobileNetV2, DenseNet121, DenseNet169, DenseNet201, NASNetMobileAbstract
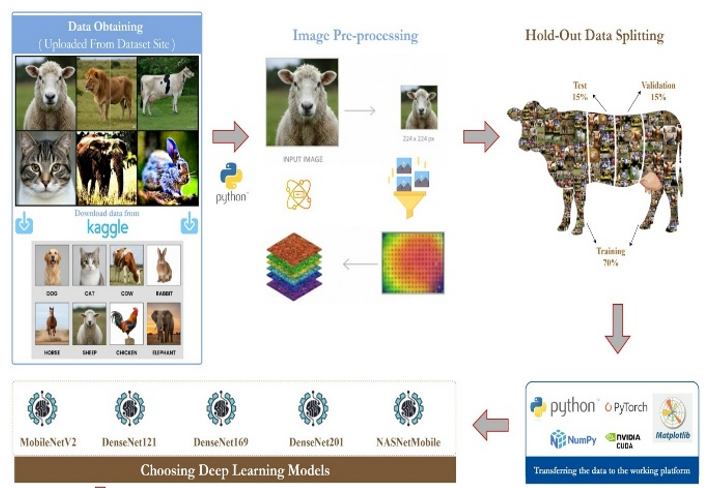
This study aims to develop a convolutional neural network (CNN)-based classification framework that can distinguish between synthetic and real animal images generated by the Stable Diffusion Turbo model. Additionally, this study will evaluate the performance of different network architectures for this task. The study employed a balanced dataset of 31,995 images, including 16,000 real images and 15,995 synthetic images generated by Stable Diffusion Turbo. The dataset includes eight animal categories: dogs, cats, cows, rabbits, horses, sheep, chickens, and elephants. All images were resized to 224 by 224 pixels, and standard preprocessing techniques were applied. During the classification stage, five pretrained convolutional neural network architectures were retrained using transfer learning, including MobileNetV2, DenseNet121, DenseNet169, DenseNet201, and NASNetMobile. Model performance was evaluated using accuracy, precision, recall, the F1 score, the area under the curve of the receiver operating characteristic, confusion matrices, and training time. The experimental results demonstrate that MobileNetV2 and DenseNet201 achieved the highest classification performance, with respective accuracy rates of 99.58% and 99.56%, and perfect area under the curve values. All DenseNet variants exhibited complete sensitivity in detecting synthetic images, whereas NASNetMobile showed substantially lower performance compared to the other models. These results suggest that synthetic images produced by diffusion-based generative models can be reliably identified when appropriately designed CNN architectures and balanced datasets are used. This provides a significant methodological contribution to the discrimination of synthetic versus real images, the detection of fake content, and the verification of visual authenticity.
Downloads
References
[1] G. Celik and M. F. Talu, "Investigation of generative adversarial network models' image generation performance," Balıkesir Üniversitesi Fen Bilimleri Enstitüsü Dergisi, vol. 22, no. 1, pp. 181-192, 2020, doi: https://doi.org/10.25092/baunfbed.679608.
[2] M. Frid-Adar, I. Diamant, E. Klang, M. Amitai, J. Goldberger, and H. Greenspan, "GAN-based synthetic medical image augmentation for increased CNN performance in liver lesion classification," Neurocomputing, vol. 321, pp. 321-331, 2018, doi: https://doi.org/10.1016/j.neucom.2018.09.013.
[3] P. Sedigh, R. Sadeghian, and M. T. Masouleh, "Generating synthetic medical images by using GAN to improve CNN performance in skin cancer classification," presented at the 2019 7th international conference on robotics and mechatronics (ICRoM), 2019. doi: 10.1109/ICRoM48714.2019.9071823.
[4] C. Han et al., "GAN-based synthetic brain MR image generation," presented at the 2018 IEEE 15th international symposium on biomedical imaging (ISBI 2018), 2018. doi: 10.1109/ISBI.2018.8363678.
[5] H. Nazki, J. Lee, S. Yoon, and D. S. Park, "Image-to-image translation with GAN for synthetic data augmentation in plant disease datasets," Smart Media Journal, vol. 8, no. 2, pp. 46-57, 2019.
[6] J. J. Bird, C. M. Barnes, L. J. Manso, A. Ekárt, and D. R. Faria, "Fruit quality and defect image classification with conditional GAN data augmentation," Scientia Horticulturae, vol. 293, p. 110684, 2022, doi: https://doi.org/10.1016/j.scienta.2021.110684.
[7] F. Wang, H. Al Hamadi, and E. Damiani, "A visualized malware detection framework with CNN and conditional GAN," presented at the 2022 IEEE International Conference on Big Data (Big Data), 2022. doi: 10.1109/BigData55660.2022.10020534.
[8] C. Dewi, R.-C. Chen, Y.-T. Liu, and S.-K. Tai, "Synthetic Data generation using DCGAN for improved traffic sign recognition," Neural Computing and Applications, vol. 34, no. 24, pp. 21465-21480, 2022, doi: https://doi.org/10.1007/s00521-021-05982-z.
[9] S. M. Sristi and M. S. Sharma, "Culturally-Aware Multiclass Bangla Text-to-Image Generation via Fine-Tuned Stable Diffusion Turbo," presented at the 2025 IEEE 11th International Conference on Big Data Computing Service and Machine Learning Applications (BigDataService), 2025. doi: 10.1109/BigDataService65758.2025.00025.
[10] A. Lokner Lađević, T. Kramberger, R. Kramberger, and D. Vlahek, "Detection of AI-generated synthetic images with a lightweight CNN," AI, vol. 5, no. 3, pp. 1575-1593, 2024, doi: https://doi.org/10.3390/ai5030076.
[11] Y. Patel et al., "An improved dense CNN architecture for deepfake image detection," IEEE Access, vol. 11, pp. 22081-22095, 2023, doi: 10.1109/ACCESS.2023.3251417.
[12] J. J. Bird and A. Lotfi, "Cifake: Image classification and explainable identification of ai-generated synthetic images," IEEE Access, vol. 12, pp. 15642-15650, 2024, doi: 10.1109/ACCESS.2024.3356122.
[13] J. Mallet, L. Pryor, R. Dave, and M. Vanamala, "Deepfake detection analyzing hybrid dataset utilizing cnn and svm," presented at the Proceedings of the 2023 7th International Conference on Intelligent Systems, Metaheuristics & Swarm Intelligence, 2023. doi: https://doi.org/10.1145/3596947.359695.
[14] A. Raza, K. Munir, and M. Almutairi, "A novel deep learning approach for deepfake image detection," Applied Sciences, vol. 12, no. 19, p. 9820, 2022, doi: https://doi.org/10.3390/app12199820.
[15] W. H. Abir et al., "Detecting deepfake images using deep learning techniques and explainable AI methods," Intelligent Automation & Soft Computing, vol. 35, no. 2, pp. 2151-2169, 2023, doi: https://doi.org/10.32604/iasc.2023.029653.
[16] H. Chen et al., "Comprehensive exploration of diffusion models in image generation: a survey," Artificial Intelligence Review, vol. 58, no. 4, p. 99, 2025, doi: https://doi.org/10.1007/s10462-025-11110-3.
[17] F. Rahat, M. S. Hossain, M. R. Ahmed, S. K. Jha, and R. Ewetz, "Data augmentation for image classification using generative ai," presented at the 2025 IEEE/CVF Winter Conference on Applications of Computer Vision (WACV), 2025. doi: 10.1109/WACV61041.2025.00410.
[18] F. Siddiqui, J. Yang, S. Xiao, and M. Fahad, "Enhanced deepfake detection with DenseNet and Cross-ViT," Expert Systems with Applications, vol. 267, p. 126150, 2025, doi: https://doi.org/10.1016/j.eswa.2024.126150.
[19] S. Banerjee, "Neural Architecture Search Based Deepfake Detection Model using YOLO," International Journal of Advanced Research in Science, Communication and Technology, pp. 375-383, 2025, doi: 10.48175/IJARSCT-22938.
[20] Y. Wu and K. He, "Group normalization," presented at the Proceedings of the European conference on computer vision (ECCV), 2018. [Online]. Available: https://link.springer.com/conference/eccv.
[21] F. He, T. Liu, and D. Tao, "Control batch size and learning rate to generalize well: Theoretical and empirical evidence," Advances in neural information processing systems, vol. 32, 2019.
[22] P. M. Radiuk, "Impact of training set batch size on the performance of convolutional neural networks for diverse datasets," 2017, doi: https://doi.org/10.1515/itms-2017-0003.
[23] M. J. Awan et al., "Image-based malware classification using VGG19 network and spatial convolutional attention," Electronics, vol. 10, no. 19, p. 2444, 2021, doi: https://doi.org/10.3390/electronics10192444.
[24] P. S. Madhyastha and R. Jain, "On model stability as a function of random seed," presented at the Proceedings of the 23rd Conference on Computational Natural Language Learning (CoNLL), 2019. doi: 10.18653/v1/K19-1087.
[25] J. Åkesson, J. Töger, and E. Heiberg, "Random effects during training: Implications for deep learning-based medical image segmentation," Computers in Biology and Medicine, vol. 180, p. 108944, 2024, doi: https://doi.org/10.1016/j.compbiomed.2024.108944.
[26] K. Dong, C. Zhou, Y. Ruan, and Y. Li, "MobileNetV2 model for image classification," presented at the 2020 2nd International Conference on Information Technology and Computer Application (ITCA), 2020. doi: 10.1109/ITCA52113.2020.00106.
[27] T. A. Cengel, B. Gencturk, E. T. Yasin, M. B. Yildiz, I. Cinar, and M. Koklu, "Automating egg damage detection for improved quality control in the food industry using deep learning," Journal of Food Science, vol. 90, no. 1, p. e17553, 2025, doi: https://doi.org/10.1111/1750-3841.17553.
[28] L. Yong, L. Ma, D. Sun, and L. Du, "Application of MobileNetV2 to waste classification," Plos one, vol. 18, no. 3, p. e0282336, 2023, doi: https://doi.org/10.1371/journal.pone.0282336.
[29] M. Eser, M. Bilgin, E. T. Yasin, and M. Koklu, "Using pretrained models in ensemble learning for date fruits multiclass classification," Journal of Food Science, vol. 90, no. 3, p. e70136, 2025, doi: https://doi.org/10.1111/1750-3841.70136.
[30] R. Indraswari, R. Rokhana, and W. Herulambang, "Melanoma image classification based on MobileNetV2 network," Procedia computer science, vol. 197, pp. 198-207, 2022, doi: https://doi.org/10.1016/j.procs.2021.12.132.
[31] Y. Ünal and M. Türkoğlu, "Mango leaf disease detection using deep feature extraction and machine learning methods: A comparative survey," El-Cezeri, vol. 12, no. 1, pp. 35-43, 2025.
[32] [32] A. Bello, S.-C. Ng, and M.-F. Leung, "Skin cancer classification using fine-tuned transfer learning of DENSENET-121," Applied Sciences, vol. 14, no. 17, p. 7707, 2024, doi: https://doi.org/10.3390/app14177707.
[33] S. A. Albelwi, "Deep architecture based on DenseNet-121 model for weather image recognition," International Journal of Advanced Computer Science and Applications, vol. 13, no. 10, 2022, doi: 10.14569/IJACSA.2022.0131065.
[34] N. A. Zebari, A. A. Alkurdi, R. B. Marqas, and M. S. Salih, "Enhancing brain tumor classification with data augmentation and densenet121," Academic Journal of Nawroz University, vol. 12, no. 4, pp. 323-334, 2023, doi: https://doi.org/10.25007/ajnu.v12n4a1985.
[35] S. Öğrekçi, Y. Ünal, and M. N. Dudak, "A comparative study of vision transformers and convolutional neural networks: sugarcane leaf diseases identification," European Food Research and Technology, vol. 249, no. 7, pp. 1833-1843, 2023.
[36] S. HARMANCI, Ü. Yavuz, and A. Barış, "Classification of hazelnut species with pre-trained deep learning models," Intelligent Methods In Engineering Sciences, vol. 2, no. 2, pp. 58-66, 2023.
[37] B. A. S. Al-Rimy, F. Saeed, M. Al-Sarem, A. M. Albarrak, and S. N. Qasem, "An adaptive early stopping technique for densenet169-based knee osteoarthritis detection model," Diagnostics, vol. 13, no. 11, p. 1903, 2023, doi: https://doi.org/10.3390/diagnostics13111903.
[38] D. Bhowmik, M. Abdullah, and M. T. Islam, "A deep face-mask detection model using DenseNet169 and image processing techniques," Brac University, 2022.
[39] P. P. Dalvi, D. R. Edla, and B. Purushothama, "Diagnosis of coronavirus disease from chest X-ray images using DenseNet-169 architecture," SN Computer Science, vol. 4, no. 3, p. 214, 2023, doi: https://doi.org/10.1007/s42979-022-01627-7.
[40] M. K. Awang et al., "Classification of Alzheimer disease using DenseNet-201 based on deep transfer learning technique," Plos one, vol. 19, no. 9, p. e0304995, 2024, doi: https://doi.org/10.1371/journal.pone.0304995.
[41] A. Dash, P. K. Sethy, and S. K. Behera, "Maize disease identification based on optimized support vector machine using deep feature of DenseNet201," Journal of Agriculture and Food Research, vol. 14, p. 100824, 2023, doi: https://doi.org/10.1016/j.jafr.2023.100824.
[42] A. Ghodekar and A. Kumar, "LightLeafNet: Lightweight and efficient NASNetmobile architecture for tomato leaf disease classification," Available at SSRN 4570347, 2023, doi: http://dx.doi.org/10.2139/ssrn.4570347.
[43] A. E. B. Alawi and A. M. Qasem, "Lightweight CNN-based models for masked face recognition," presented at the 2021 International Congress of Advanced Technology and Engineering (ICOTEN), 2021. doi: 10.1109/ICOTEN52080.2021.9493424.
[44] H. Güven and A. Saygılı, "Monkeypox Diagnosis Using MRMR-Based Feature Selection and Hybrid Deep Learning Models: ResNet50V2, NASNetMobile, and InceptionV3," International Scientific and Vocational Studies Journal, vol. 9, no. 1, pp. 173-182, 2025, doi: https://doi.org/10.47897/bilmes.1706322.
[45] A. W. A. Ameer, P. Salehpour, and M. Asadpour, "Deep transfer learning for lip reading based on NASNetMobile pretrained model in wild dataset," IEEE Access, 2024, doi: 10.1109/ACCESS.2024.3470521.
[46] I. A. Ozkan and M. Koklu, "Skin lesion classification using machine learning algorithms," International Journal of Intelligent Systems and Applications in Engineering, vol. 5, no. 4, pp. 285-289, 2017, doi: 10.18201/ijisae.2017534420.
[47] Y. Karatas et al., "Identification of Leaf Diseases from Figs Using Deep Learning Methods," Selcuk Journal of Agriculture and Food Sciences, vol. 38, no. 3, pp. 414-426, 2024, doi: 10.15316/SJAFS.2024.037.
[48] E. Yasin and M. Koklu, "A comparative analysis of machine learning algorithms for waste classification: inceptionv3 and chi-square features," International Journal of Environmental Science and Technology, vol. 22, no. 10, pp. 9415-9428, 2025, doi: https://doi.org/10.1007/s13762-024-06233-z.
[49] R. Kursun and M. Koklu, "Classification of Eggplant Diseases Using Feature Extraction with AlexNet and Random Forest," presented at the 4th International Conference on ContemporaryAcademic Research, Konya, Turkey, February 22-23, 2025, 2025.
[50] H. Incekara, I. H. Cizmeci, M. M. Saritas, and M. Koklu, "Classification of almond kernels with optuna hyper-parameter optimization using machine learning methods," Journal of Food Science and Technology, pp. 1-17, 2025, doi: https://doi.org/10.1007/s13197-025-06494-7.
[51] T. A. Cengel, B. Gencturk, E. T. Yasin, M. B. Yildiz, I. Cinar, and M. Koklu, "Apple (malus domestica) quality evaluation based on analysis of features using machine learning techniques," Applied Fruit Science, vol. 66, no. 6, pp. 2123-2133, 2024, doi: https://doi.org/10.1007/s10341-024-01196-4.
[52] M. Koklu, R. Kursun, E. T. Yasin, and Y. S. Taspinar, "Detection of Defects in Soybean Seeds by Extracting Deep Features with SqueezeNet," presented at the 2023 IEEE 12th International Conference on Intelligent Data Acquisition and Advanced Computing Systems: Technology and Applications (IDAACS), 2023. doi: 10.1109/IDAACS58523.2023.10348939.
[53] M. C. Hinojosa Lee, J. Braet, and J. Springael, "Performance metrics for multilabel emotion classification: comparing micro, macro, and weighted f1-scores," Applied Sciences, vol. 14, no. 21, p. 9863, 2024, doi: https://doi.org/10.3390/app14219863.
[54] M. Koklu and I. A. Ozkan, "Multiclass classification of dry beans using computer vision and machine learning techniques," Computers and Electronics in Agriculture, vol. 174, p. 105507, 2020, doi: https://doi.org/10.1016/j.compag.2020.105507.
[55] M. Koklu, I. Cinar, and Y. S. Taspinar, "Classification of rice varieties with deep learning methods," Computers and electronics in agriculture, vol. 187, p. 106285, 2021, doi: https://doi.org/10.1016/j.compag.2021.106285.
[56] Y. S. Taspinar, I. Cinar, R. Kursun, and M. Koklu, "Monkeypox Skin Lesion Detection with Deep Learning Models and Development of Its Mobile Application," presented at the International Journal of Research in Engineering and Science (IJRES), January 2024, 2024.
[57] Y. S. Taspinar, I. Cinar, and M. Koklu, "Prediction of computer type using benchmark scores of hardware units," presented at the Selcuk University Journal of Engineering Sciences April 2021, 2021.
Downloads
Published
Issue
Section
License
Copyright (c) 2025 Intelligent Methods In Engineering Sciences

This work is licensed under a Creative Commons Attribution-ShareAlike 4.0 International License.






