Deep Learning-Based Classification of Skin Lesion Dermoscopic Images for Melanoma Diagnosis
DOI:
https://doi.org/10.58190/imiens.2024.101Keywords:
Deep learning, Lesion Classification, Melanoma, Skin Cancer, Skin LesionAbstract
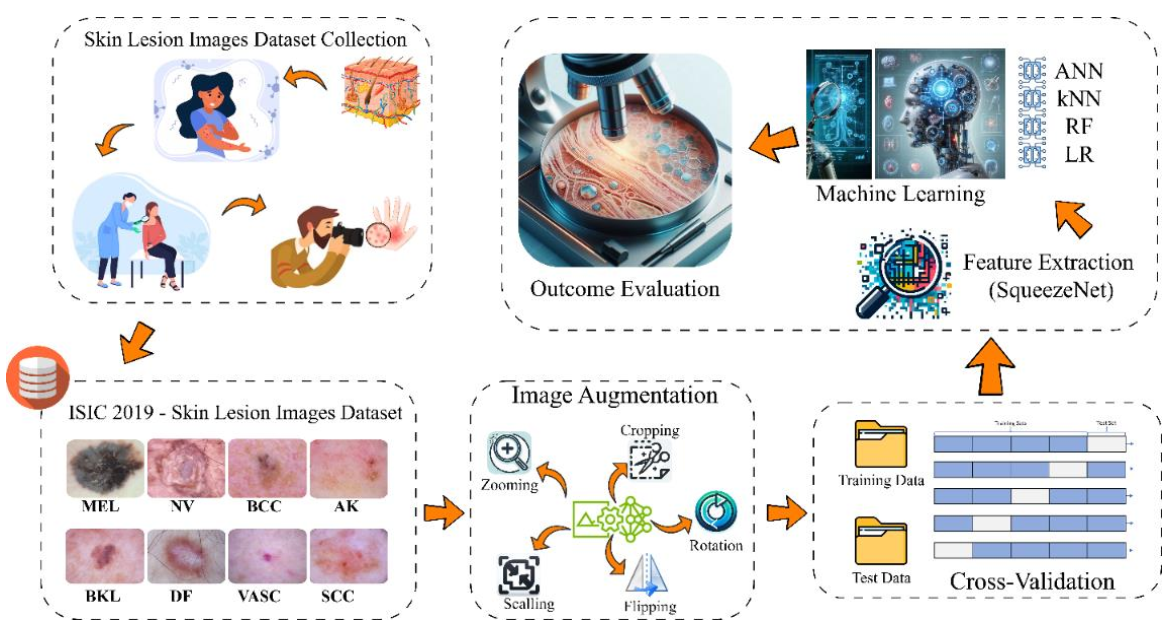
This study examined the effectiveness of artificial intelligence and machine learning methods in accurately categorizing skin cancers. We used the ISIC 2019 Skin Lesion dataset, which consists of images divided into eight categories. To prepare the data, we applied preprocessing techniques and extracted features using the SqueezeNet deep learning model. The dataset was divided into training and test sets using cross validation. Four well-known machine learning algorithms, namely Artificial Neural Network (ANN), k-nearest neighbors (kNN), Random Forest (RF), and Logistic Regression (LR), were employed to perform classification tasks. Each algorithm was specifically designed to suit its processing methodology. The algorithms are assessed based on several important measures. The results indicate that the Artificial Neural Network (ANN) achieved the highest accuracy rate of 71.80%, whereas the k-nearest neighbors (kNN), Random Forest (RF), and Logistic Regression (LR) achieved accuracy rates of 69.70%, 67.00%, and 67.20%, respectively. The results emphasize the capacity of machine learning algorithms to augment clinical decision-making in the field of dermatology, with the goal of enhancing early detection and treatment effectiveness for individuals with skin cancer.
Downloads
References
K. Safdar, S. Akbar, and S. Gull, "An automated deep learning based ensemble approach for malignant melanoma detection using dermoscopy images," in 2021 International Conference on Frontiers of Information Technology (FIT), 2021: IEEE, pp. 206-211.
K. Feng, L. Ren, G. Wang, H. Wang, and Y. Li, "SLT-Net: A codec network for skin lesion segmentation," Computers in Biology and Medicine, vol. 148, p. 105942, 2022.
X. Wang, X. Jiang, H. Ding, Y. Zhao, and J. Liu, "Knowledge-aware deep framework for collaborative skin lesion segmentation and melanoma recognition," Pattern Recognition, vol. 120, p. 108075, 2021.
A. Mahbod, P. Tschandl, G. Langs, R. Ecker, and I. Ellinger, "The effects of skin lesion segmentation on the performance of dermatoscopic image classification," Computer Methods and Programs in Biomedicine, vol. 197, p. 105725, 2020.
P. Choudhary, J. Singhai, and J. Yadav, "Skin lesion detection based on deep neural networks," Chemometrics and Intelligent Laboratory Systems, vol. 230, p. 104659, 2022.
N. S. A. ALEnezi, "A method of skin disease detection using image processing and machine learning," Procedia Computer Science, vol. 163, pp. 85-92, 2019.
S. Pathan, K. G. Prabhu, and P. Siddalingaswamy, "Techniques and algorithms for computer aided diagnosis of pigmented skin lesions—A review," Biomedical Signal Processing and Control, vol. 39, pp. 237-262, 2018.
A. Uong and L. I. Zon, "Melanocytes in development and cancer," Journal of cellular physiology, vol. 222, no. 1, pp. 38-41, 2010.
Y. S. Taspinar, I. Cinar, R. KURSUN, and M. KOKLU, "Monkeypox Skin Lesion Detection with Deep Learning Models and Development of Its Mobile Application," Public health, vol. 500, p. 5.
H. W. Rogers, M. A. Weinstock, S. R. Feldman, and B. M. Coldiron, "Incidence estimate of nonmelanoma skin cancer (keratinocyte carcinomas) in the US population, 2012," JAMA dermatology, vol. 151, no. 10, pp. 1081-1086, 2015.
J. Feng, N. G. Isern, S. D. Burton, and J. Z. Hu, "Studies of secondary melanoma on C57BL/6J mouse liver using 1H NMR metabolomics," Metabolites, vol. 3, no. 4, pp. 1011-1035, 2013.
H. Li, Y. Pan, J. Zhao, and L. Zhang, "Skin disease diagnosis with deep learning: a review," Neurocomputing, vol. 464, pp. 364-393, 2021.
P. Tschandl, C. Rosendahl, and H. Kittler, "The HAM10000 dataset, a large collection of multi-source dermatoscopic images of common pigmented skin lesions," Scientific data, vol. 5, no. 1, pp. 1-9, 2018, doi: 10.1038/sdata.2018.161.
M. Arif, F. M. Philip, F. Ajesh, D. Izdrui, M. D. Craciun, and O. Geman, "Automated detection of nonmelanoma skin cancer based on deep convolutional neural network," Journal of Healthcare Engineering, vol. 2022, 2022.
D. Gavrilov, A. V. Melerzanov, N. Shchelkunov, and E. Zakirov, "Use of neural network-based deep learning techniques for the diagnostics of skin diseases," Biomedical Engineering, vol. 52, pp. 348-352, 2019.
Z. Waheed, A. Waheed, M. Zafar, and F. Riaz, "An efficient machine learning approach for the detection of melanoma using dermoscopic images," in 2017 International conference on communication, computing and digital systems (C-CODE), 2017: IEEE, pp. 316-319.
A. A. Adegun and S. Viriri, "Deep learning-based system for automatic melanoma detection," IEEE Access, vol. 8, pp. 7160-7172, 2019.
S. Serte and H. Demirel, "Gabor wavelet-based deep learning for skin lesion classification," Computers in biology and medicine, vol. 113, p. 103423, 2019.
M. K. Hasan, L. Dahal, P. N. Samarakoon, F. I. Tushar, and R. Martí, "DSNet: Automatic dermoscopic skin lesion segmentation," Computers in biology and medicine, vol. 120, p. 103738, 2020.
S. N. Hasan, M. Gezer, R. A. Azeez, and S. Gulsecen, "Skin lesion segmentation by using deep learning techniques," in 2019 Medical Technologies Congress (TIPTEKNO), 2019: IEEE, pp. 1-4, doi: 10.1109/TIPTEKNO.2019.8895078.
P. Shan, Y. Wang, C. Fu, W. Song, and J. Chen, "Automatic skin lesion segmentation based on FC-DPN," Computers in Biology and Medicine, vol. 123, p. 103762, 2020.
A. Murugan, S. A. H. Nair, A. A. P. Preethi, and K. S. Kumar, "Diagnosis of skin cancer using machine learning techniques," Microprocessors and Microsystems, vol. 81, p. 103727, 2021.
D. d. A. Rodrigues, R. F. Ivo, S. C. Satapathy, S. Wang, J. Hemanth, and P. P. Reboucas Filho, "A new approach for classification skin lesion based on transfer learning, deep learning, and IoT system," Pattern Recognition Letters, vol. 136, pp. 8-15, 2020.
N. Nida, A. Irtaza, A. Javed, M. H. Yousaf, and M. T. Mahmood, "Melanoma lesion detection and segmentation using deep region based convolutional neural network and fuzzy C-means clustering," International journal of medical informatics, vol. 124, pp. 37-48, 2019.
M. K. Monika, N. A. Vignesh, C. U. Kumari, M. Kumar, and E. L. Lydia, "Skin cancer detection and classification using machine learning," Materials Today: Proceedings, vol. 33, pp. 4266-4270, 2020.
I. A. Ozkan and M. Koklu, "Skin lesion classification using machine learning algorithms," International Journal of Intelligent Systems and Applications in Engineering, vol. 5, no. 4, pp. 285-289, 2017, doi: 10.18201/ijisae.2017534420.
Y. S. Taspinar, I. Cinar, and M. Koklu, "Classification by a stacking model using CNN features for COVID-19 infection diagnosis," Journal of X-ray science and technology, vol. 30, no. 1, pp. 73-88, 2022, doi: 10.3233/XST-211031.
M. Combalia et al. BCN_20000, doi: 10.48550/arXiv.1908.022.
N. C. Codella et al., "Skin lesion analysis toward melanoma detection: A challenge at the 2017 international symposium on biomedical imaging (isbi), hosted by the international skin imaging collaboration (isic)," in 2018 IEEE 15th international symposium on biomedical imaging (ISBI 2018), 2018: IEEE, pp. 168-172, doi: 10.48550/arXiv.1710.05006.
N. Codella et al., "Skin lesion analysis toward melanoma detection 2018: A challenge hosted by the international skin imaging collaboration (isic)," arXiv preprint arXiv:1902.03368, 2019, doi: 10.48550/arXiv.1902.03368.
E. Avuclu, A. Elen, and H. Kahramanli Ornek, "Making inferences about settlements from satellite images using glowworm swarm optimization," Journal of Electrical Engineering & Technology, vol. 15, pp. 2345-2360, 2020, doi: 10.1007/s42835-020-00509-3.
R. Kursun, I. Cinar, Y. S. Taspinar, and M. Koklu, "Flower recognition system with optimized features for deep features," in 2022 11th Mediterranean Conference on Embedded Computing (MECO), 2022: IEEE, pp. 1-4, doi: 10.1109/MECO55406.2022.9797103.
M. Kizilgul et al., "Real-time detection of acromegaly from facial images with artificial intelligence," European journal of endocrinology, vol. 188, no. 1, pp. 158-165, 2023, doi: 10.1093/ejendo/lvad005.
I. Cinar, "Disaster Detection Using Machine Learning Methods with Deep Features," disasters, vol. 19, p. 21, 2023.
R. Butuner, I. Cinar, Y. S. Taspinar, R. Kursun, M. H. Calp, and M. Koklu, "Classification of deep image features of lentil varieties with machine learning techniques," European Food Research and Technology, vol. 249, no. 5, pp. 1303-1316, 2023, doi: 10.1007/s00217-023-04214-z.
Y. Unal, Y. S. Taspinar, I. Cinar, R. Kursun, and M. Koklu, "Application of pre-trained deep convolutional neural networks for coffee beans species detection," Food Analytical Methods, vol. 15, no. 12, pp. 3232-3243, 2022, doi: 10.1007/s12161-022-02362-8.
I. Cinar, Y. S. Taspinar, and M. Koklu, "Development of early stage diabetes prediction model based on stacking approach," Tehnički glasnik, vol. 17, no. 2, pp. 153-159, 2023, doi: 10.31803/tg-20211119133806.
Y. S. Taspinar, "Diabetic Rethinopathy phase identification with deep features," in 2022 11th Mediterranean conference on embedded computing (MECO), 2022: IEEE, pp. 1-4, doi: 10.1109/MECO55406.2022.9797136.
F. Bre, J. M. Gimenez, and V. D. Fachinotti, "Prediction of wind pressure coefficients on building surfaces using artificial neural networks," Energy and Buildings, vol. 158, pp. 1429-1441, 2018, doi: 10.1016/j.enbuild.2017.11.045.
G. Guo, H. Wang, D. Bell, Y. Bi, and K. Greer, "KNN model-based approach in classification," in On The Move to Meaningful Internet Systems 2003: CoopIS, DOA, and ODBASE: OTM Confederated International Conferences, CoopIS, DOA, and ODBASE 2003, Catania, Sicily, Italy, November 3-7, 2003. Proceedings, 2003: Springer, pp. 986-996.
R. Gandhi, "Nearest Neighbours—Introduction to Machine Learning Algorithms," Towards Data Science, 2018.
G. Biau and E. Scornet, "A random forest guided tour," Test, vol. 25, pp. 197-227, 2016.
D. G. Kleinbaum, K. Dietz, M. Gail, M. Klein, and M. Klein, Logistic regression. Springer, 2002.
E. T. Yasin, I. A. Ozkan, and M. Koklu, "Detection of fish freshness using artificial intelligence methods," European Food Research and Technology, vol. 249, no. 8, pp. 1979-1990, 2023, doi: 10.1007/s00217-023-04271-4.
K. Erdem, M. B. Yildiz, E. T. Yasin, and M. Koklu, "A Detailed Analysis of Detecting Heart Diseases Using Artificial Intelligence Methods," Intelligent Methods In Engineering Sciences, vol. 2, no. 4, pp. 115-124, 2023, doi: 10.58190/imiens.2023.71.
A. Feyzioglu and Y. S. Taspinar, "Malicious UAVs classification using various CNN architectures features and machine learning algorithms," International Journal of 3D Printing Technologies and Digital Industry, vol. 7, no. 2, pp. 277-285, 2023, doi: 10.46519/ij3dptdi.1268605.
B. Gencturk et al., "Detection of hazelnut varieties and development of mobile application with CNN data fusion feature reduction-based models," European food research and technology, vol. 250, no. 1, pp. 97-110, 2024, doi: 10.1007/s00217-023-04369-9.
E. Abdullayeva and H. K. Ornek, "Diagnosing Epilepsy from EEG Using Machine Learning and Welch Spectral Analysis," Traitement du Signal, vol. 41, no. 2, 2024, doi: 10.18280/ts.410237.
S. B. Khojasteh and H. K. Ornek, "The Robust EEG Based Emotion Recognition using Deep Neural Network," International Journal of Intelligent Systems and Applications in Engineering, vol. 9, no. 4, pp. 191-197, 2021, doi: 10.18201/ijisae.2021473639.
Downloads
Published
Issue
Section
License
Copyright (c) 2024 Intelligent Methods In Engineering Sciences

This work is licensed under a Creative Commons Attribution-ShareAlike 4.0 International License.






